Common User Issues
1) I cannot find my haplogroup.See Enter SNP
2) The sample on the map is not indicative of the exact place I trace my descent.
HRAS only has access to the country and regional codes visible on the public YFull YTree page.
In the future there will be an integration with YSEQ customers' uniparental male and female line self-reported origins, exact to the latitude longitude they enter.
3) Some haplogroup centroids are computed over the water.
The haplogroup centroid is the haplogroup centroid, computed by an algorithm that takes sample geographic coordinates and their position in the Y / mtDNA tree into account. It is a relatively simple, deterministic (same inputs always result in same outputs) algorithm (not powered by AI) that does not take water or land terrain features into account.
See Centroids
DNA type
Click to switch between Y and mtDNA analysis modes.Map type
Clicking a different map type will reset the map to that map type and load the current haplogroup.Original Documentation of each map type:
Relative Frequency Heatmap (the page says mtDNA but methodology applies to both)
Relative Frequency Heatmap (Classic)
Diversity Heatmap
Relative Frequency (Classic) and Diversity Heatmaps use the Leaflet heatmap.js plugin designed by Patrick Wied.
Relative Frequency Heatmap uses my own software for computing and rendering the relative frequency and is more accurate for our specific use case (it divides a target frequency surface by a denominator frequency surface for each point on the map), especially in revealing regional substructure.
You may notice some differences between the original classic and diversity heatmaps and how they appear in HRAS.
- The controls have been streamlined.
- The surfaces may look different for the same haplogroup the radius is now configurable.
- The relative frequency estimates in classic may be slightly different because the denominators come from the new codebase (computed in js rather than python) and the legend is calibrated with the more accurate relative frequencies computed by the newer heatmap version.
- The orginal diversity map always used the same radius for each sample. The HRAS version applies radius and intensity to each sample based on regional code area according to the same logic for each map type as defined in section Sample Minimum Radius.
Enter haplogroup or SNP
Start typing the haplogroup or SNP, then click the desired subclade from the autocompleted results.Only those haplogroups downstream of supported haplogroup roots are supported by HRAS.
Only the SNPs defining these haplogroups from the YFull YTree or MTree are supported.
Y-DNA root haplogroups: A00, A0, A1a, A1b1, B, C, D, E, F-Y27277, G, H, I1, I2, J1, J2, K-Y28299, L, M, N, O, Q, R1a, R1b, R2, S, T
mtDNA root haplogroups: DN, NA, U, R-a, R9c, R9b, F, R8, R7, R6, R5, R4, R32, R31, R30, R3, R2, T, J, R23, R22, R14, R13, R12'21, R10, R1, R0a'b, HV-b, HV-a, HV5, HV4, HV32, HV31, HV30, HV3, HV29, HV28, HV27, HV26, HV25, HV21, HV19, HV18, HV13, HV12, HV1, HV0k, HV0j, HV0i, HV0h, V, HV0a7, HV0a6, HV0a5, HV0a4, HV0a3, HV0a2, HV0a1, HV0-a, H, P, A, N10, N11, N13, N14, N1a1a, I, N1a1b1, N1a2, N1a3, N1b, N1c, N5, N2a, W, N21, N22, N3, N6, N7, N8, N9a, N9b, Y, ND, L0, L1, L2, L3a, L3b'f, L3c'd, L3e'i'k'x, L3h, M10, M11, M1'20'51, G, M12, M13'46'61, M14, M15, M16, M17, M19'53, M2, M21, M22, M23'75, M24'41, M25, M26, M27, M28, M29, M29'Q1, Q, M3, M31, M32'56, M33, M34'57, M35, M36, M39'70, M40, M42'74, M44, M4''67, M47, M48, M49, M5, M50, M52, M55'77, M58, M59, M6, M62'68, M69, M7, M71, M72, M73'79, M76, M78, C, Z, M8a, D, M80, M80'D1, M81, M82, M83, M84, M85, M86, E, M9a'b, M91, O, S, X, L4, L5'7, L6
Ancient vs Modern Samples
HRAS provides an ancient / modern toggle to show the changes in relative frequency of a haplogroup between various ancient time intervals and today.It should be noted that the reliability of the computed relative frequency of ancient samples will be lower than that of modern samples, due to having less overall samples and greater holes in coverage, i.e. areas where no ancient samples whatsoever have yet been recovered.
Samples

Samples representing the same regional code are arranged in concentric circles around the center. The example above shows the J-Z2507 samples with geocodes for Albania, not specifying a more precise region within Albania.
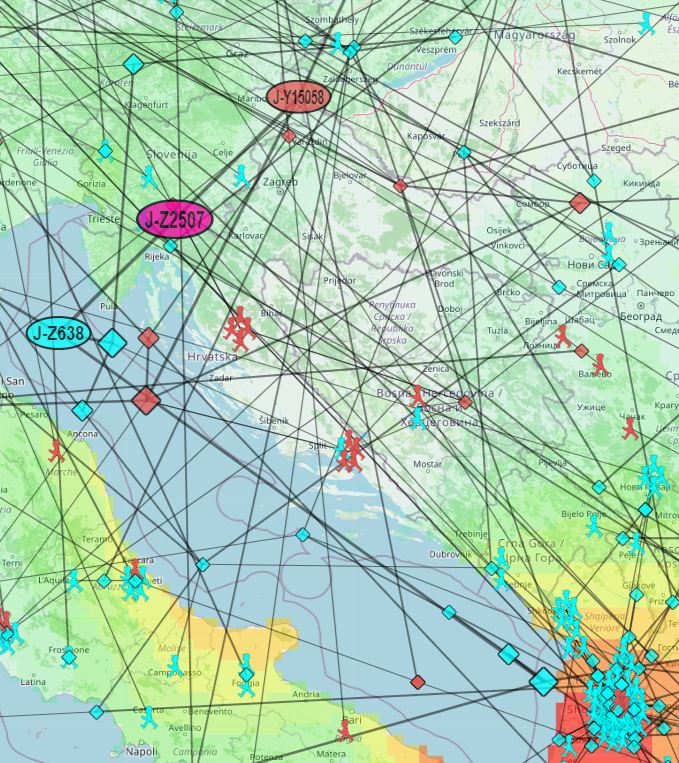
The color of the sample corresponds to the color assigned to the subclade in the "Migration from Target to Children" view.
Country Statistics
This layer adds a vertically aligned pie slice over each country center containing samples of the target haplogroup.The radius of the pie slice indicates total number of samples for that country, regardless of their haplogroup.
The angle measure of the pie slice indicates the fraction of samples that are positive for the target haplogroup.
Clicking the country adds a popup showing a pie chart.
Statistics are computed based on the modern samples that trace their uniparental descent to / ancient samples that were found within a given country's modern borders.
What is displayed differs between ancient and modern sampling modes.
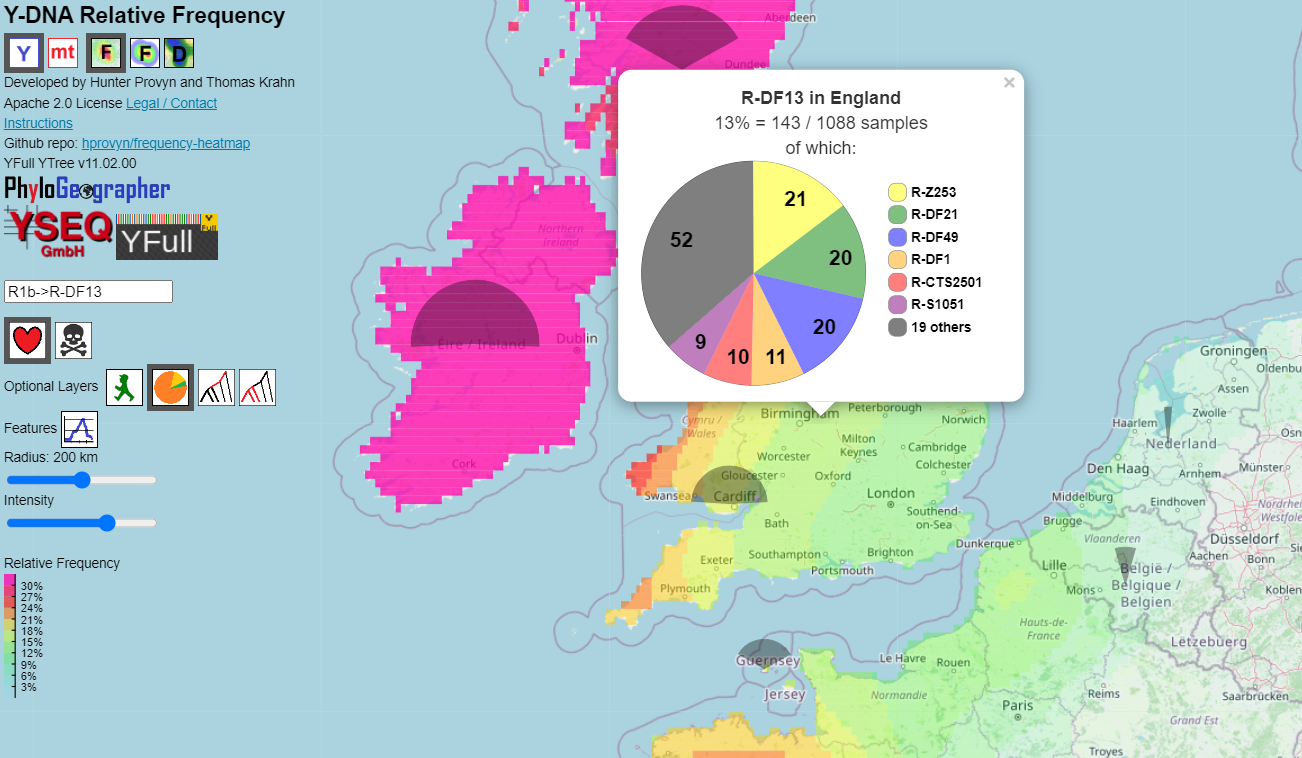
For modern, the whole pie represents all modern samples from that country that are positive for the haplogroup.
The sections indicate the number and proportion of samples in various downstream lineages.
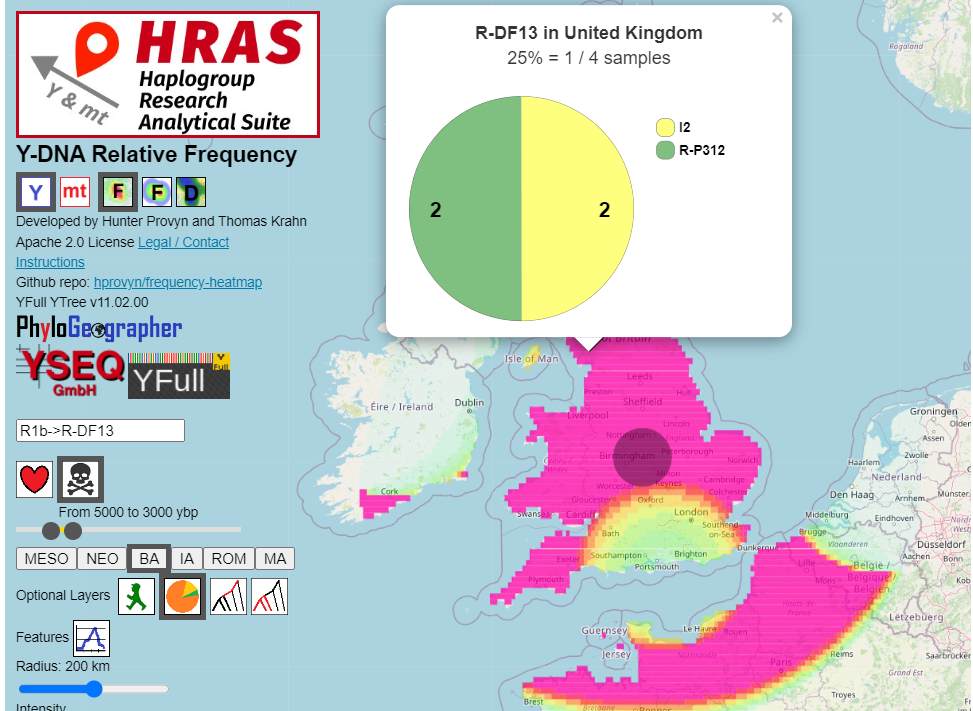
For ancient, the whole pie represents all ancient samples found within that country whose sample age falls within the specified time interval.
The sections indicate the number and proportion of ancient samples in each of various major haplogroups that YFull designates as [haplogroup roots].
Animation
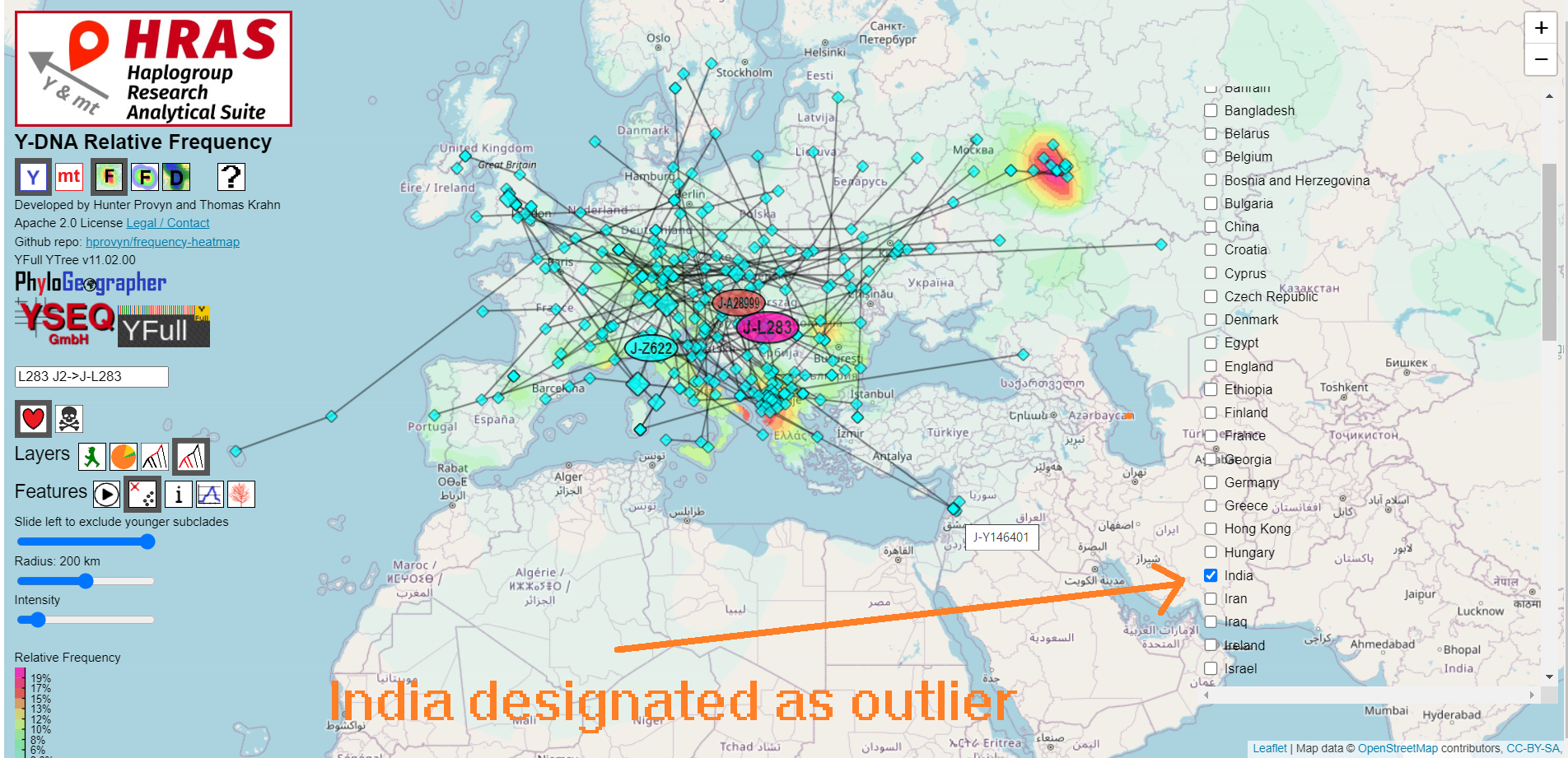
Clicking the animation button gradually slides the TMRCA slider from left to right, slowly adding to the map the samples and/or subclades computed centroids (depending on which are enabled) in the order of their subclade's TMRCA.Outliers
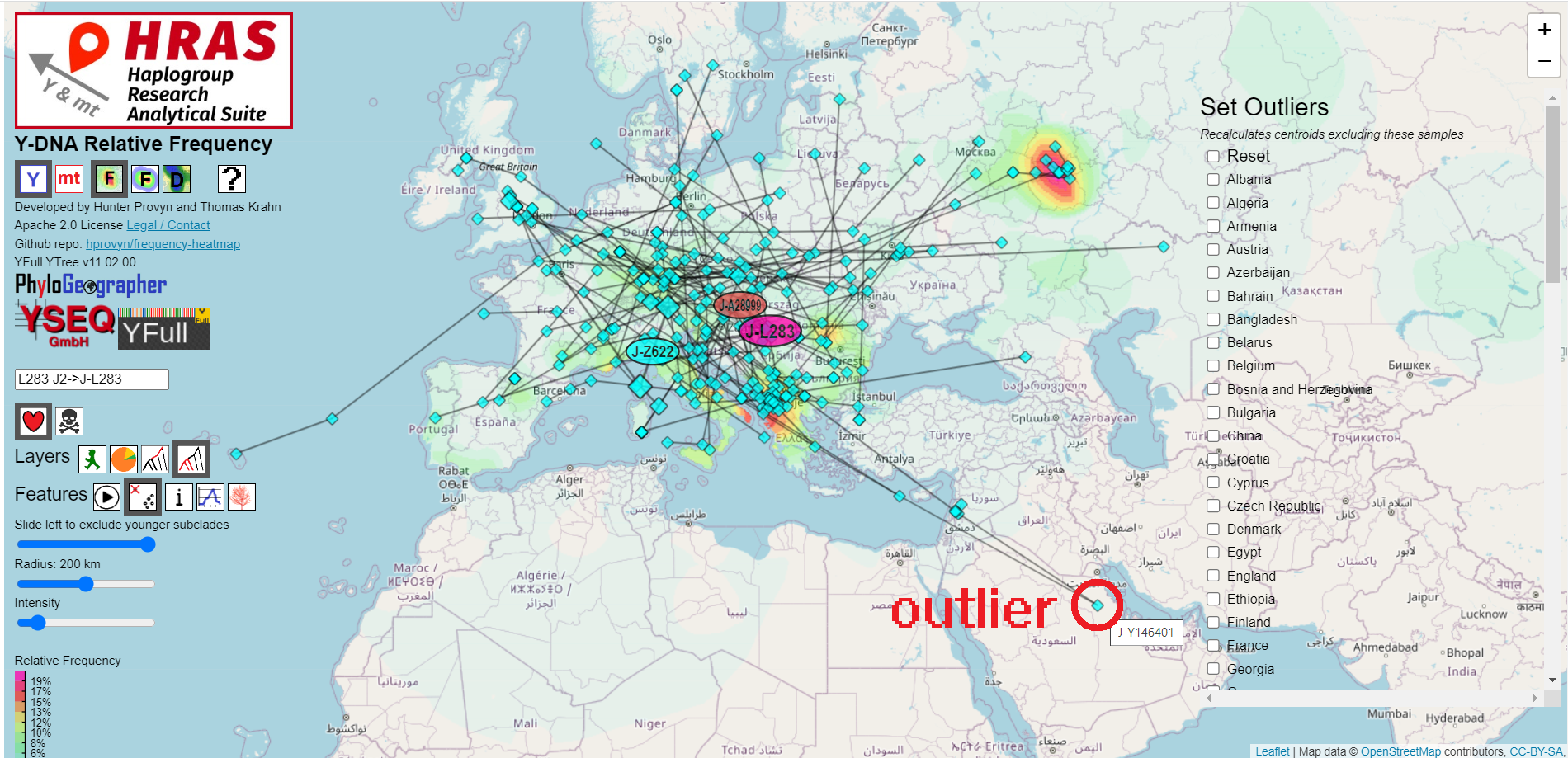
HRAS does not automatically determine whether samples from a certain region should be ignored on the basis of being extreme outliers.You may specify which countries to exclude via the "Outliers" control.
Centroids
The centroid is a representation of the approximate geographic center of the haplogroup as computed by the regional codes of the self-reported ancestry locations of samples and their position in the tree below the target haplogroup.For a technical description of the algorithm Centroid Computation Algorithm
Centroid Metrics

For each child subclade of the target haplogroup, Centroid Metrics displays the average distance from its children nodes to its computed centroid, along with other information.
A child node of a haplogroup is either a child subclade, a basal sample, or a regular sample in the case that the haplogroup has no child subclades.
"Elapsed Time" is the estimated elapsed time between the target haplogroup's TMRCA and the TMRCA of the relevant subclade.
"Mean Migration Rate" is computed by dividing the distance by elapsed time. Keep in mind that these figures are not to be taken as the absolute truth regarding the migration, but they are objectively computed by inputs.
Theory
Lower nodes-to-centroid distance is an indicator (read: not a guarantee) of higher reliability that the computed centroid represents the approximate geographic origin of that subclade's MRCA.You could consider the possibility that the MRCA was living in that approximate area.
The child nodes there are, the more reliable the computed centroid.
However, the reliability should be called into question if you doubt the reliability of the computed centroids of any of the child nodes.
Or if an ancient sample that is nearly as old as the subclade's TMRCA was found in a completely different and impossibly distant region.
However, in that case the centroid computation algorithm should favor the older ancient sample proportional to its age.
Migration from Haplogroup Root to Target
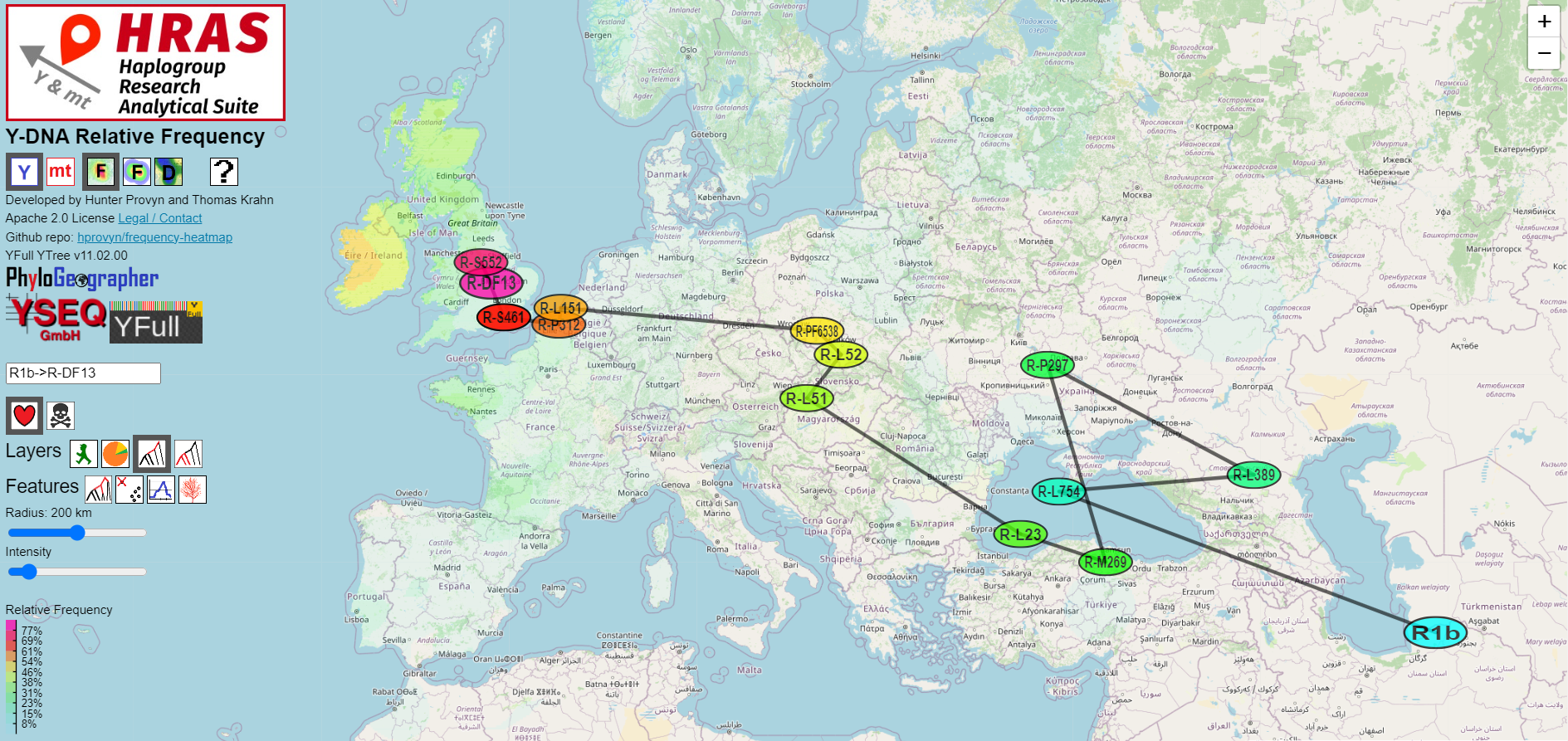
HRAS connects the centroids of subsequent branches with lines, producing an approximate migration path across the map.
Keep in mind that several factors make it unrealistic to expect that an algorithm with this kind of input data can 100% reliably compute the true migration path:
- discrepancies in regional sampling rates
- lack or paucity of ancient samples
- errors in self-reported ancestral locations
- lack of geographic consistency of self-reported ancestral locations
- the ever-present possibility of mass migration from a homeland where all men in the original location either moved in the same direction or died out
Keeping this in mind, researchers should consider the HRAS-computed centroids as just one bit of information that should be taken into consideration as part of a multidisciplinary-approach with other data - including samples from other sources and less readily quantifiable types of information from, but not limited to the archeaological, historical or climatological record.
Migrations from Target to Children
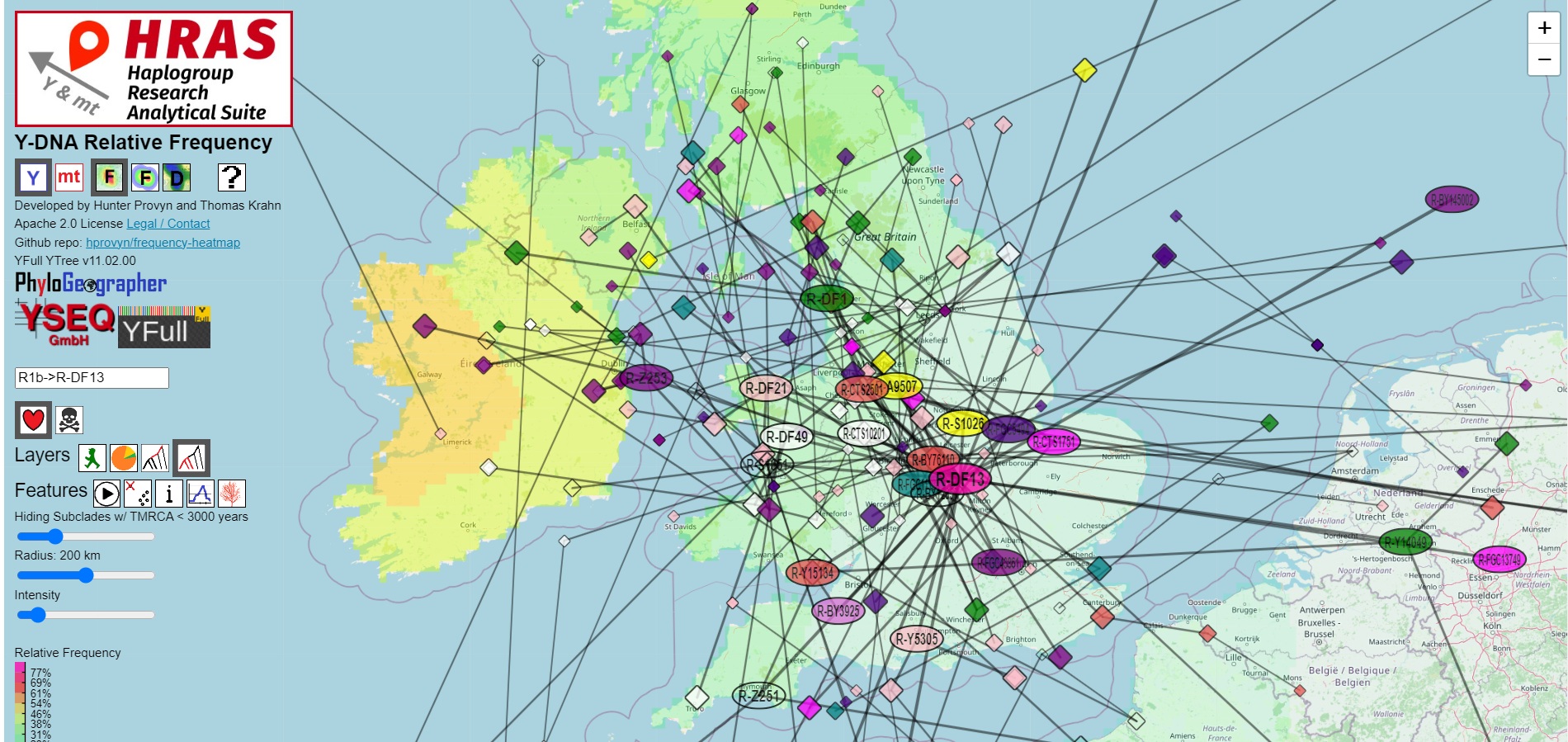
HRAS connects the centroids of subsequent branches with lines, producing an approximate migration path across the map.
Large and medium diamonds represent the immediate child and grandchildren subclades of the labeled subclades.
Smaller diamonds are used to represent all other downstream subclades.
Keep in mind that several factors make it unrealistic to expect that an algorithm with this kind of input data can 100% reliably compute the true migration path:
- discrepancies in regional sampling rates
- lack or paucity of ancient samples
- errors in self-reported ancestral locations
- lack of geographic consistency of self-reported ancestral locations
- the ever-present possibility of mass migration from a homeland where all men in the original location either moved in the same direction or died out
Keeping this in mind, researchers should consider the HRAS-computed centroids as just one bit of information that should be taken into consideration as part of a multidisciplinary-approach with other data - including samples from other sources and less readily quantifiable types of information from, but not limited to the archeaological, historical or climatological record.
Diversification Timeline

The raw data used to compute the diversification over time graph are counts of how many lineages formed (using YFull nomenclature) during every 100 year interval going back in time to the target haplogroup's estimated TMRCA.
These raw counts are then smoothed using binomial coefficients [1,6,15,20,15,6,1]. So the smoothed figure for the number of lineages representing the hundred year interval 2500 years ago would be:
1/64 * 2800 ybp count + 6/64 * 2700 ybp count + 15/64 * 2600 ybp count + 20/64 * 2500 ybp count + 15/64 * 2400 ybp count + 6/64 * 2300 ybp count + 1/64 * 2200 ybp count
This graph can be exported as a SVG by clicking the button.
TMRCA Time Slider
Use this slider to hide samples and computed centroids for younger (by TMRCA) haplogroups.You may want to use this to declutter the map of more recently formed geographic outliers or founder effects.
Sample Minimum Radius
The minimum radius is the radius used in the heatmap for samples representing regions with less than 10,000 km2 area.Samples represented by regional codes of larger area have correspondingly longer radius, but the intensity is reduced so that a sample always contributes the same overall weight on the map.
Increasing the minimum radius has the effect of smoothing the surface at the cost of losing any regional differentiation.
Technical
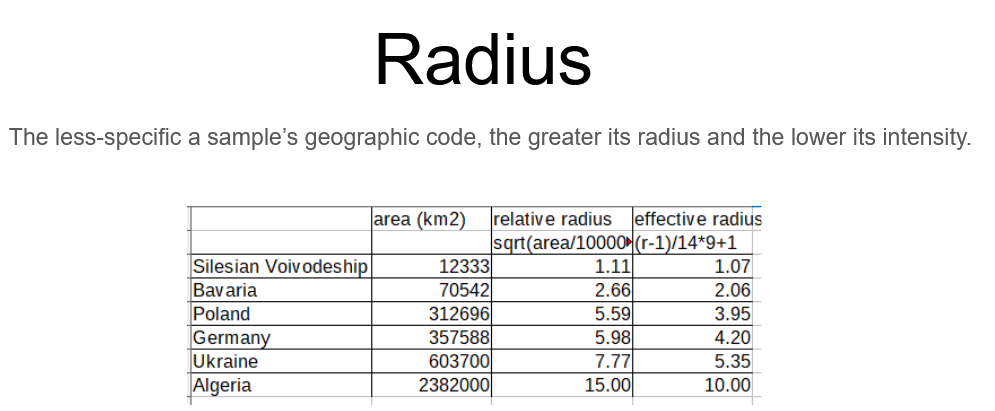
Two steps to get the effective minimum radius used by HRAS:
- Divide the area of the geographic code's region by 10,000 km2 and then take the square root to get a number between 1-15
- Translate this number to a 1-10 scale
Intensity
By adjusting this slider you can increase or decrease the intensity of the heatmap.For the relative frequency heatmap, the legend will update accordingly.
The diversity heatmap legend does not change because it is unitless.
Future
Geodesic Lines
Use Geodesic lines to connect centroids to represent approximate migration paths. This will look better and be more accurate than the standard leaflet line in showing the shortest path between two points on the Earth. I found a leaflet plugin I can use.Migration Animation
I plan to implement a similar functionality as PhyloGeographer that uses interpolation to gradually trace the path between connected centroids, from the haplogroup root to the target haplogroup.YSEQ Samples
HRAS will integrate samples tested at YSEQ that have specified uniparental male or female line geographic origins through the YSEQ interface.Research/Project Links
I envision adding functionality to HRAS that will facilitate sharing of pertinent information by link that may be set by a haplogroup / surname clan researcher.This could include links to projects and HRAS parameters (i.e. country outliers) / views that the researcher feels conveys useful information for someone viewing the target haplogroup.
Some features may require a subscription fee on behalf of the researcher (not regular user) to HRAS to support the maintenence and development of new features.
